Friday, September 30, 2016
A Newbie at Kamino
As everyone knows, I have been interning with Kathleen at TGen. She is a really awesome person from Wisconsin and I am now starting to believe all really cool people come from Wisconsin. These past couple of weeks I have been learning about CRISPR, and I have been learning the TGen SDS system. This past week I was in the lab helping Kathleen with sorting out reagents from kits to dispose. We had to compartmentalize everything based on their pictograms and chemicals. It is such a great experience working in the TGen office.
S-STEM 4th Semester at Kamino
Hello everyone!
My name is Cel Parra and this will be my fourth semester in the SSTEM program. If you have seen my previous blog posts, I have worked with ant DNA and cactus DNA. I specifically researched how closely the DNA was related to each other. I researched how closely related the ants were by observing the DNA bands to determine whether they came from the same nest. I did the same with DNA from cholla cactus spines to determine whether they reproduce sexually or asexually. This Fall 2016 term I am interning at the main TGen building downtown with Kathleen Kennedy. She is the Manager of Environmental Health & Safety. It is going really well, and I cannot wait to see what the rest of this semester holds for me at TGen!
Thursday, May 5, 2016
The Continuation of Spines from Tatooine
Over the course of my time left in SSTEM, I still plan on
continuing my research of cholla spines. I hope to sequence my bands in the
future at ASU, and I hope to have some concrete results about Cylindropuntia bigelovii reproduction. I
now have a solid idea of what method to use and what primers work best for the teddy-bear
cholla DNA. I have found some new prospective people to work with, and their additional
help to my project will help further guide me to the results I hope to gain from this research. This summer is going to be pretty hectic, but I cannot wait
to return back in the fall to poke myself with these spines again.
Thursday, April 28, 2016
EMCC Presentation of Tusken Spines
Today I did an oral presentation on the work I have been
doing in the lab. I presented the best method, in my experiment, to extract DNA
from cholla spines. The best method for the extractions was by using the ZYMO™
Plant DNA kit. The best primers to use were the Universal Rice primers because they
actually amplified the DNA, and I was able to see banding. It was a good experience and my last at an EMCC conference. Now I am on to
creating a poster for the final presentations at Metro Tech. I will be going
with a tan outline of my poster and not so much blue. I hope it does not hurt
anyone’s feelings here at Phoenix College.
Thursday, April 21, 2016
The Sand People Emerge
This week I was able to determine what were the best methods in going about extracting DNA from the cholla spines I collected. Below are results from prickly pear cactus spines and how their banding looked after two extraction methods, but both extraction methods ran with the same Universal Rice Primers. I cannot wait to exert these methods upon the teddy-bear cholla spines collected from Dreamy Draw.
These are the gels from the first DNA extraction from the kit ran with Universal Rice Primers 1-4:
These are the gels from the first DNA extraction with InstaGene matrix ran with Universal Rice Primers 1-4:
These are the gels from the first DNA extraction from the kit ran with Universal Rice Primers 1-4:
These are the gels from the first DNA extraction with InstaGene matrix ran with Universal Rice Primers 1-4:
As you can see, both DNA samples are amplified, but the better banding results from the first extraction method.
Thursday, April 7, 2016
Collecting Tusken Spines from Tusken heads
The day right before spring break was a blast! My mentors
and my SSTEM peer Lucas traveled through Dreamy Draw for a nice, early hike. We
saw some pretty interesting insects, including a Gila monster. We found some
nice Cylindropuntia bigelovii
populations and we were able to collect some spines for my DNA extraction. I
wish more people had gone to experience what a great time we had. I was pretty
worn out by the end of the trip and wanted to crash out. It was all worth it
though since I had gotten the spines I needed.
Here is a video of us finding the Gila monster:
Thursday, March 31, 2016
Another PCR Run with Tusken Raider Spines
This week I ran another PCR with my DNA samples with the same chloroplast primers. The first PCR run included 10 microliters of distilled water, 10 microliters DNA samples, and 20 microliters of the plant master mix that included the chloroplast primers. The only changes to the protocol was increasing the amount of water to 15 microliters and changing the amount of DNA sample to 5 microliters. The amount of plant master mix into the PCR tube stayed the same. The idea was for the primers to have a higher chance of targeting the small amount of DNA to increase amplification. The result of this PCR after electrophoresis is presented below in this gel:
We assumed the ladder ran off a bit weird due to how the gels were made. I'll need to run another fresh gel with my PCR samples. For the mean time, I'll be working on amplifying the DNA samples with other primers in the lab that include primers 1-7.
We assumed the ladder ran off a bit weird due to how the gels were made. I'll need to run another fresh gel with my PCR samples. For the mean time, I'll be working on amplifying the DNA samples with other primers in the lab that include primers 1-7.
Thursday, March 24, 2016
Harvesting from Multiple Tusken Raiders & Amplification
Before spring break, I was able to collect spines from a population of teddy-bear cholla (cylindropuntia bigelovii) at the dreamy draw hike. Below are some pictures of the population of cholla cacti, and both of my mentors got in my way.
I ran a PCR on my five samples with chloroplast primers and the results were not what I expected. Since I did not know what kind of primers were in the master mix, I did not know which primer to avoid when running a PCR with the
DNA samples. The next step was to run a RAPD PCR with primers we have in the lab to see if the amplification would be better than the amplification from the Chloroplast primers. Below is a gel of the DNA samples amplified with the chloroplast primers.
Here is also a video of one of our top highlights at the Dreamy Draw hike:
Thursday, March 10, 2016
The Future of the Tusken Raider Spines
This week I was able to run a gel on five samples of DNA I have extracted. The most recent extraction was of prickly pear spines for which I changed the protocol by the time I conducted the extraction. The sample was cleaned in twine and bleach before it was heated for three hours at 70 degrees Celsius. I ran the sample along with four others, including the control, but I was not able to see any bands. The control sample of Trifolium sp. had a band in the first lane, but the rest of the cactus spine samples did not. The next step is to run a PCR and I hope to see DNA banding after the PCR samples undergo electrophoresis. Here is my gel below:
Thursday, March 3, 2016
Spines from a Tusken Raider's Head
This week I worked on extracting DNA from prickly pear spines in hopes of perfecting a protocol in order to see some banding in my gels. I was able to collect only 40 mg of cholla spines, so Josh was able to collect some prickly pear spines for me in his front yard. Below is a picture of how much 40 mg of cholla spines looks like. It is not very much and I need a packed tube of cholla spines to conduct multiple extractions of 150 mg each.
The difference in this protocol I conducted this week was how there was a 3-hour incubation period at seventy degrees Celsius. It seemed as though this worked because the spines looked like they had turned to a pulp and there would be a more likely chance of extracting DNA. I also used small scissors to cut up the pieces and did some grinding. After running the gel though, I did not see any DNA bands. The next suggestion, given to me by Matt, was to grind the spines with a pestle and mortar to create a powdered substance from the spines and use 150 mg of that plant material for my next extraction. I am hoping it will work.
Thursday, February 25, 2016
Looking up to Endor
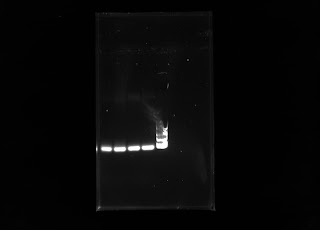
This week I was able to run some gels on the two cholla DNA extractions I had completed last week. Unfortunately, I did not see any bands after running electrophoresis. To make sure I was conducting the correct steps from the protocol, I did a DNA extraction on a trifolium sp. and was able to run electrophoresis on the sample. It turns out I was doing the protocol correctly, since there were bands in the gel! Here is the gel:
Polymorphic banding of Trifolium sp.
In two of the lanes from the wells, we are able to see two bright bands of DNA from the Trifolium sp.! It's exciting and I hope to see some bands in my next extraction from spines.
Thursday, February 4, 2016
Finding Parts from Jakku
This week I completed another extraction with the ZR Plant/Seed DNA MiniPrep™ kit, but included an overnight incubation in room temperature and an extra 10 minutes in a man-mad bead beater. Josh was able to come up with a new way of bashing my beads by creating an awesome "home-made" bead beater here in the lab. Choosing between buying an actual bead beater or creating one, Josh's superior sagacity thought up of creating one. I was able to bash my beads without the numbness taking over my hands and the beating of the beads would be consistent. I saw improvement in the bashing of the cholla spines and the solution actually turned a bit yellow. This did not happen previously with my first extraction and I am hoping it was a serious improvement. I'll see if there are any bands next week after the samples undergo electrophoresis.
Thursday, January 28, 2016
The Awakening Begins
https://www.zymoresearch.com/dna/microbial-environmental-dna-isolation-1/soil-fecal-plant-dna/zr-plant-seed-dna-miniprep
I will be conducting this experiment carefully because I do not want to end up looking like this.
Subscribe to:
Posts (Atom)